The second edition of training on Next Generation Sequencing (NGS) of SARS-CoV-2 started on December 1 at ACEGID, Redeemer’s University, Ede, Nigeria. The training is aimed at strengthening genomic surveillance capacity on the African continent.
Following opening formalities, Dr Yenew Kebede, Head of Laboratory Systems at Africa Centres for Disease Control and Prevention (Africa CDC) and Prof. Christian Happi, Director ACEGID gave remarks, projecting the vision behind the training as well as expectations from the participants.
“This training is very relevant to genomic surveillance because as we are sharing knowledge and empowering other countries that do not have those skills. They go back and start doing genomic surveillance for their countries. Then, they can be ahead of the curve; we can become more proactive instead of being responsive the way we are right now,” said Prof Happi.
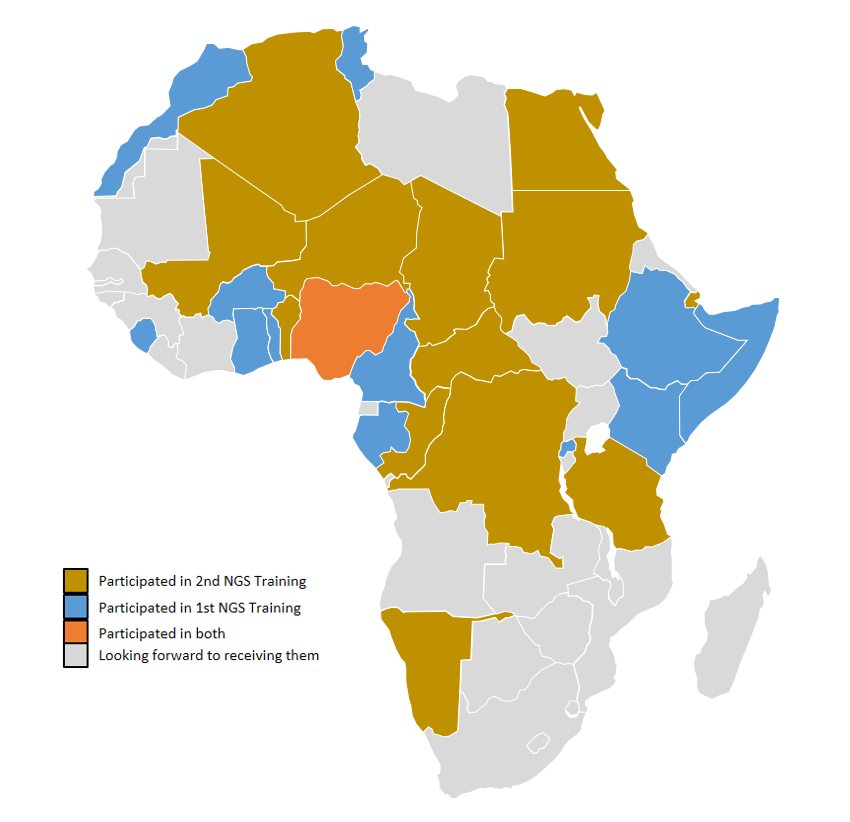
The two-weeks training will have scientists from 13 African countries in attendance. The participants are involved in national SARS-CoV-2 sequencing efforts in their respective countries. Participants will have hands-on training on the entire sequencing process: from extraction of RNA through to sequencing and bioinformatics analysis.
The participants came with high expectations from the training. “My expectation is to be able to know everything about next generation sequencing, starting from different methods and ending with data analysis,” said Mawahib Eldegail, Specialist in NGS Lab at National Public Health Laboratory, Khartoum, Sudan.
Musuamba Pauline, Biologist Medical (Medical Biologist) at Institut National de la Recherche Biomédicale, Kinshasha, Congo, would like to “master the technique of viral whole genome sequencing.”

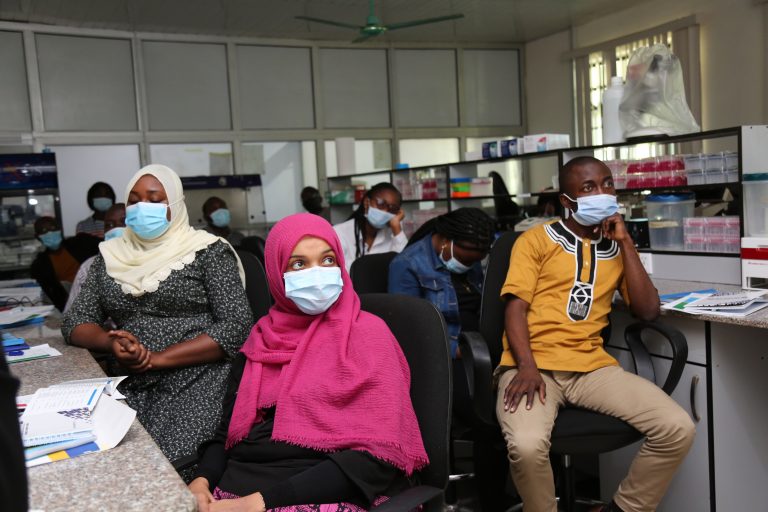
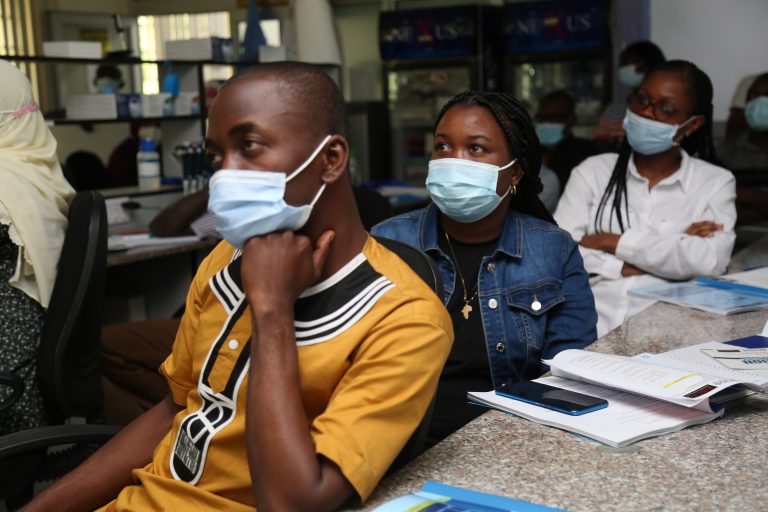


Participants will be exposed to different tools, protocols and platforms that are available for sequencing SARS-CoV-2 and for analysing the genome data.
Mahdi Elmi, Laboratory Supervisor at the Molecular Biology National Laboratory, Ministry of Health, Balbala, Djibouti, hopes to “learn more about sequencing platforms, especially the Oxford Nanopore Technology MinION and Illumina Miseq for Covid-19.”
Participants at the training will gain the capabilities to independently sequence, analyze and share SARS-CoV-2 data. They will also be able to interpret SARS-CoV-2 sequence data, and to use this data as evidence to inform public health response.
“This training gives ACEGID the opportunity to propagate the knowledge, therefore, democratizing NGS technology; by training others who, in turn, will become trainers in their countries,” said Prof. Happi. “It is a Training of Trainers that we are having here.”
This training is part of ACEGID’s effort to develop genomics capacity across Africa. The first edition of the NGS Training had scientists from 14 African countries in attendance. Earlier this week, our team concluded a two-week technical support training on SARS-CoV-2 Genome Sequencing and Bioinformatics Analysis for scientists at The Institut Pasteur de Côte d’Ivoire, Abidjan, Côte d’Ivoire.
The training is organized in partnership with Africa Centres for Disease Control and Prevention (Africa CDC), African Society for Laboratory Medicine (ASLM) and World Health Organization Africa Region, with sponsorship from Illumina and Redeemer’s University.